Drug Discovery Online Assessment
- Country :
Australia
Drug Discovery Online Tutorial 3
Part A: Finding a druggable pocket
In VMD, open up the protein structure of PA2G4. HINT: same structure used in the proteins structure online tutorial i.e. pdb code: 2q8k-2.pdb
Change the Drawing Method until you have a view where you can visualise the surface of the protein. HINT: CPK, surf etc. Also try typing in the selected atoms box not protein and not water.
- Can you find any potential druggable pockets? HINT: deep cavities in the protein. Take a screenshot and add to your report.
- There are other molecules crystallised with this protein, what are they? Are these druggable pockets?
PA2G4 is thought to interact with the oncogene MYCN.
- Would this protein-protein interaction site be expected to be on the surface of PA2G4, or buried?
Now open up the structure PA2G4_MYCN.pdb.
- Change the drawing method and colouring method until you can clearly see the 2 separate proteins. Take a snapshot and add to the report.
- Why is the full protein structure of MYCN not shown? HINT: What do we know about MYC?
- Find 3 interactions between MYC and PA2G4? HINT: Polar interactions, hydrophobic.
Take a snapshot of each and describe the type and distance between the two atoms
Part B: Conducting and analysing a Virtual Screen
This site will now be used to conduct a simple computational Virtual Screen using the program EasyVS.
Upload the PDB file 2q8k-2.pdb. Click next step (Step 2). The program will now identify potential druggable pockets.
- How many pockets does it find?
- How many of these are considereddruggable?
Choose Pocket 2 (same as the MYCN binding pocket) and click next step (Step 3). Click the button to Set to RO3. Select DrugBank. Reduce Similarity to 0.1. Click Process library. HINT: Time Estimation should be around 9 minutes. Click Next Step (Step 4).
- What was the top hit? Take a screenshot and add to the report. NB: This may hang, just take a picture of the top hit even if job isnt finished.
- What are the pharmacokinetic features of this compound and are these druglike?
- Open up the excel spreadsheet VS_results.xlsx This file obtains the top 45 drugs that were obtained from a large Virtual Screen with scoring and physicochemical properties added. Select the top 10 compounds to purchase. List these in the table below. HINT: Which are going to be the best hit compounds for a drug discovery campaign?
Part C: Developing a Drug
Go back to VMD and open up the structure PA2G4_drug.pdb. Change the drawing method until you have a view where you can visualise the ligand (drug) and the protein. HINT: Try typing in not protein to find the drug.
- Take a screenshot which highlights how the ligand and protein interact with each other. How do you think this drug functions i.e does it stop an endogenous ligand from binding? Does it stop another protein from binding? Etc.
- Find 2 interactions between the ligand and the protein. HINT: Could be polar interactions, covalent, hydrophobic.
Take a snapshot of the 2 interactions and describe the type and distance between the two atoms.
- Look at how the drug interacts with the protein. Can you spot any areas where you could grow the drug? Take a snapshot of these and add to this report.
The following drug is a known inhibitor of PA2G4
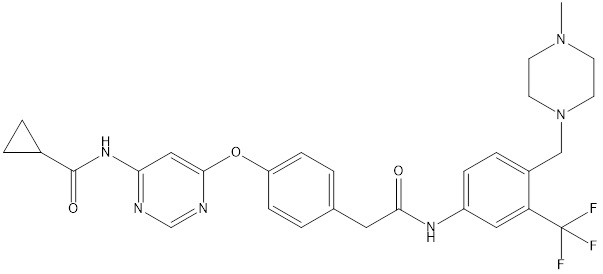
Go to "http://biosig.unimelb.edu.au/pkcsm/prediction"http://biosig.unimelb.edu.au/pkcsm/prediction
Input the following SMILES string (HINT: copy and paste into the box)
O=C(C1CC1)NC2=NC=NC(OC3=CC=C(CC(NC4=CC=C(CN5CCN(C)CC5)C(C(F)(F)F)=C4)=O)C=C3)=C2
Input the following SMILES string and calculate the toxicity (HINT: you can either choose ADMET, or just the toxicity button)
O=C(C1CC1)NC2=NC=NC(OC3=CC=C(CC)C=C3)=C2
- Is this smaller compound expected to be as toxic?
- Circle the part of the compound you predict to be causing the hERG II inhibition